EDUCATION
1995-2001
|
Department of Chemistry, Nankai University,Tianjin, China.
Received Ph.D. degree in June, 2001 (with Prof. Yun-Ti Chen).
|
1991-1995
|
Department of Chemistry, Qufu Normal University, Qufu,
China. Received B. Sc. Degree in June, 1995.
|
ACADEMIC CAREER
l PhD (with Professor Chen Yun-Ti), Nankai University (2001)
l Postdoctoral (with Professor Ivano Bertini), Center of Magnetic Resonance, University of Florence, Florence, Italy (2001-2004)
l Research fellow (with Professor Gottfried Otting), Australian National University, Canberra, Australia (2004-2010)
|
长期从事化学生物学、生物无机化学和生物核磁共振方法学研究,已在国外核心期刊上发表研究论文
50
余篇,
H-index 28.
目前课题组主要研究方向是生物大分子的定点修饰化学和高分辨生物核磁共振方法学,发展研究离体和在体环境下大分子的动态结构与作用机制的高分辨磁共振方法,并利用该技术阐明大分子的作用机制、聚集行为与功能的关系;顺磁核磁共振研究弱相互作用下主客体分子识别机制与小分子化合物结构测定。
研究方向:化学生物学、镧系离子与结构生物学、生命有机分析化学
Research Interests: Chemical biology, Bioanalytical chemistry, NMR spectroscopy
We are interested in site-specific modifications of proteins both in vitro and in cells, and use these tagging methods to characterize the structures, interactions and dynamics of proteins in vitro and in cells with NMR, EPR and other biophysical techniques.
研究内容
1)有机合成化学:功能有机小分子探针的设计与合成;非天然氨基酸的设计与合成;生物大分子标记化学与应用:生物大分子标记策略与标记中的化学反应性
2)小分子/大分子-大分子作用的高分辨分析方法:小分子与大分子作用的高分辨结构;
3)有效药物先导分子的筛选与优化
4)蛋白质动态学研究的新方法与新技术
具体为:蛋白质上的化学反应性;蛋白质定点标记技术和应用;功能蛋白质原位研究技术和方法;铜离子的稳态平衡与细胞内动态行为控制机制;酶催化过程的动态机制与底物识别;Ras蛋白细胞膜瞬态作用的分子机制与功能
基金资助
1. 国家自然科学基金面上项目(21073101 ):“用于结构生物学和药物研发的蛋白质顺磁标记”, 2011. 1-2013. 12, 主持
2. 国家自然科学基金面上项目(21273121 ):“位点特异标记稀土金属离子在蛋白质构象分析中的应用”, 2013.1 – 2016.12,主持
3. 国家重大研究计划973项目(2013CB910200):“蛋白质动态学研究的新技术、新方法”, 2013. 1 - 2017. 8,参与
4. 国家自然科学基金面上项目(21473095):“利用顺磁核磁共振研究固有无归结构蛋白质的探索”, 2015.1-2018.12, 主持
5. 国家自然科学基金面上项目(21673122):“利用赝接触位移研究蛋白质瞬态复合物三维结构的测定方法”, 2017.1-2020.12,主持
6. 国家重点研发计划(2016YFA0501202):”蛋白质机器的动态结构的核磁共振研究方法及应用”, 2016.7-2021.6,参与
7. 重点国际合作基金(中以合作)(21761142004):“发展应用于细胞内蛋白质结构、动态变化的顺磁探针与磁共振方法”,2017.10-2020.9,主持
邀请报告
2017. 3. 27-28, “3D structure determination of low-abundance and unstable enzyme intermediate using PCS”, 生物磁共振研讨会,中国科学技术大学,合肥
2017. 6. 11-17,”3D Structure Determination of Unstable Transient Enzyme Intermediates Using PCSs”, 戈登会议-生物核磁计算分会(GRC-Biomolecular NMR),美国,缅因州,Sunday River
2017. 7. 22-28, “Stable paramagentic tags in analysis of structures and dynamics of proteins in vitro and in cells using paramagnetic NMR spectroscopy”, 国际磁共振会议 (ISMAR),加拿大魁北克
2017. 9. 22-24,”3D structure determination of low-abundance and unstable enzyme intermediate using PCS”, 第六届全国“跨学科蛋白质研究”学术讨论会,广州
2017. 10. 9-11,”3D structure determination of proteins in living cells using paramagnetic NMR spectroscopy”, 第十七届北京分析测试学术报告会(BCEIA2017),北京
2017. 10. 19-22, “高性能顺磁探针在测定不稳定蛋白质复合物三维结构与细胞内环境研究中的应用”, 第二届全国生物磁共振研讨会,青岛
2017.11.3-6, “Stable paramagnetic tags in analysis of structures and dynamics of proteins in vitro and in cells using NMR and EPR”, 第十七届生物物理大会,上海
在读研究生
王晓,杨峰,陈甚娜,张凌阳,公言军,潘彬彬,李尚,赵丽娜,翟程梁,肖宇昊,李夏妍
本科生
方恬,王文举,于正鑫,赵阳,温志辉
已毕业研究生
李庆峰,魏珍,黄凤,杨茵,王金涛,左会会,裴莹莹,马飞贺,刘洪开,张琳琳,赵宇,侯萌萌,曹婵,李骘,陈家良
已毕业本科生
金科,吉佳,夏君,何龙
We have been focusing on chemical modifications of proteins with functional groups especially for NMR and EPR analysis both in vitro and in living cells. Using this technique, we attempt to delineate the dynamics, structures and interactions of protein and protein and protein-ligand interactions. With developed tagging methods, molecular recognition and dynamics at atomic resolution in cells will be ultimately discovered.
Research Highlights
Site-specific labeling of proteins with lanthanide ions for high-resolution NMR in structural biology and chemical biology
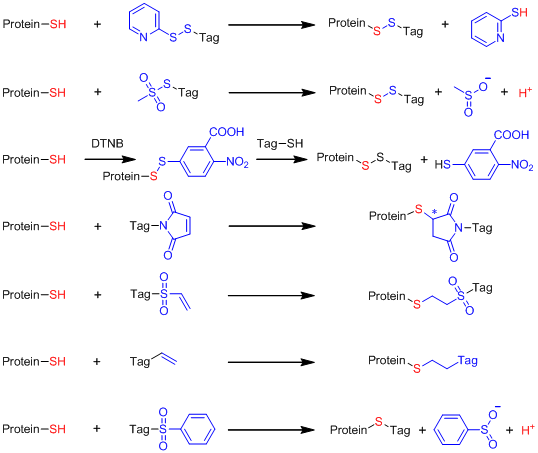
Strateg* in site-specific labeling of proteins with paramagnetic species for NMR and EPR study.
1. Site specific labeling of proteins with a paramagnetic tag
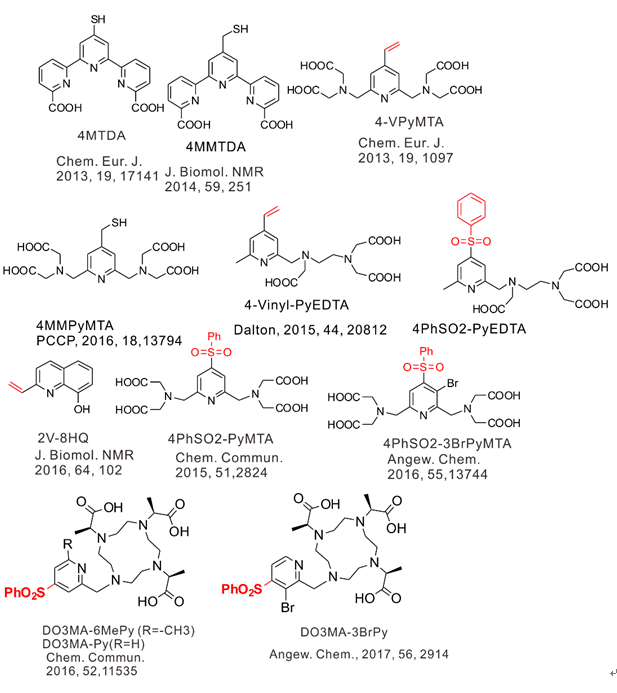
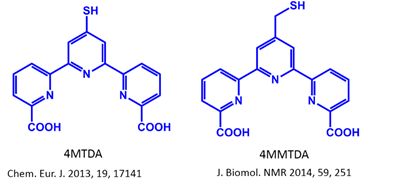
1) Labeling of proteins with dual functional tags for protein assay: 4MTDA and 4MMTDA are rigid fluorescent and paramagnetic tags, which can be attached to protein with the formation of a disulfide bonds
(Chem Eur J 2013, 17141 and J Biomol NMR 2014, 59, 251).
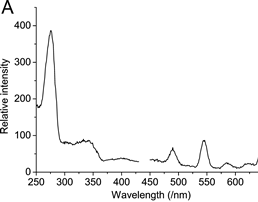
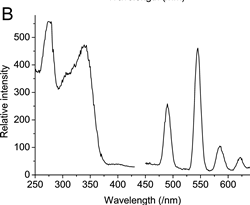
UV and fluorescence spectra of Tb(III) complexed with 4MTDA tagged ubiquitin at different sites.
2) Site specific labeling of proteins with Michael addition like thio-ene reaction: mild condition in aqueous solution (pH about 7.6-9), no generation of new chiral center, and no radical initiation
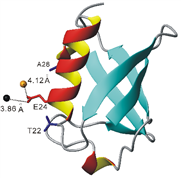
(Chem Comm 2012, 2704, Eur J Chem 2013, 1097 and J Biomol NMR 2014, 251).

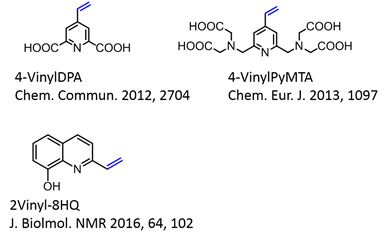
4-VinylPyMTA forms stable complex with lanthanide ions and has shown to be suitable for in situ NMR analysis (Chem Eur J 2013, 1097) and then was further applied for in-cell EPR by Drescher et al in JACS 2014, 136, 15366.
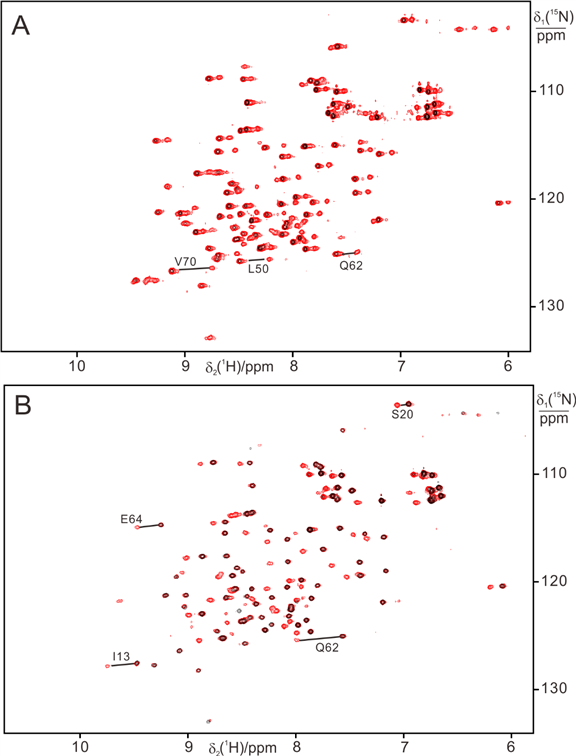
15N-HSQC indicated lanthanide complex with 4-Vinyl PyMTA conjugated protein is stable in high concentrations of HEWL (300 mg) (A) and BSA (100 mg) (B).
The protein-2V-8HQ forms a stable complex with transition metal ions, Co(II), Cu(II), Mn(II) and Ni(II), and the paramagnetic effects generated by these paramagnetic ions were evaluated by NMR spectroscopy. We show that 2V-8HQ is a rigid and stable transition metal binding tag, of which the coordination to metal ion is assisted by protein sidechain. Tunable paramagnetic tensors can be simply achieved in an α helix that possesses a i and i+4 segment, where i depicts residue Glu(or His) and i+4 the residue to be mutated to cysteine, respectively. Extensive PRE effects have been evaluated. Reliable PREs can be achieve by the high-binding affinity of paramagnetic tag.
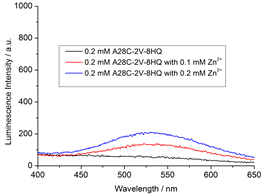
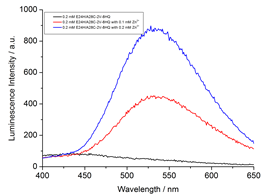
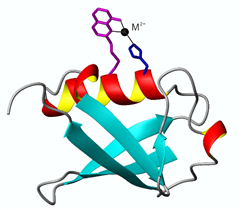
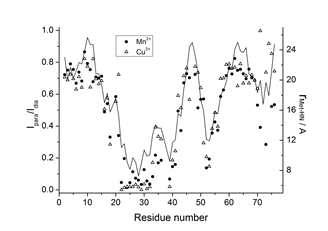
The paramagnetic anisotropy and fluorescence of protein-2V-8HQ conjugate with metal complex can be tuned by the coordination of amino acid sidechains in the helix.
3) Phenylsulfonated pyridine derivatives are rigid and stable lanthanide binding tags.
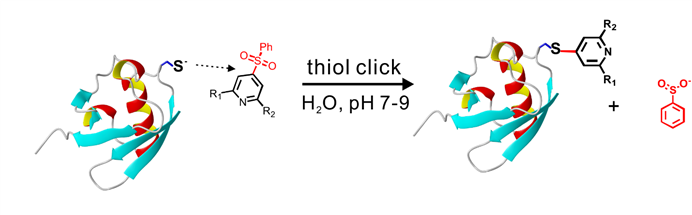
High stability of phenylsulfonated pyridine derivatives permits the in-cell NMR and EPR measurements.
4) Selective 15N-labeling of Gln and Asn sidechains and combination with paramagnetic tagging
We developed an efficient way of selectively labeling the side chains of asparagine, or asparagine and glutamine residues with 15NH2.
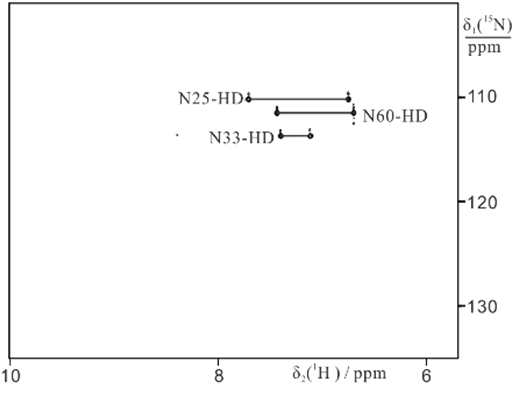
15NH2-Asn labeled ubiquitin
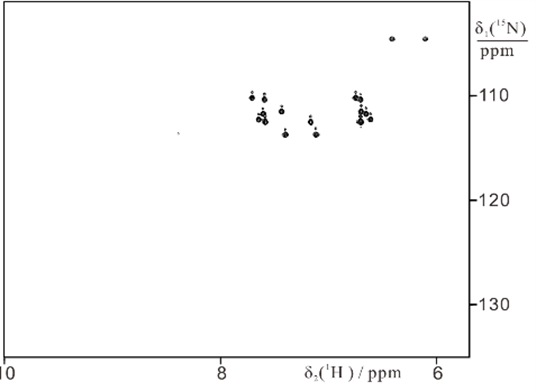
15NH2-Asn + Gln labeled ubiquitin
We developed an efficient way of selectively labeling the side chains of asparagine, or asparagine and glutamine residues with 15NH2. Those side-chains lend themselves particularly
well to determinations of Dc tensors, offering a promising approach to study protein–ligand complexes of high-molecular weight systems where the signals of side-chain amides are more easily
detected than the resonances of backbone amides.
Application of paramagnetic tagging proteins in structural biology.
1) Protein dynamics delineated by paramagnetic NMR spectroscopy: multi-domain replacement (in collaboration with Professor Gottfried Otting and Professor Thomas Huber)
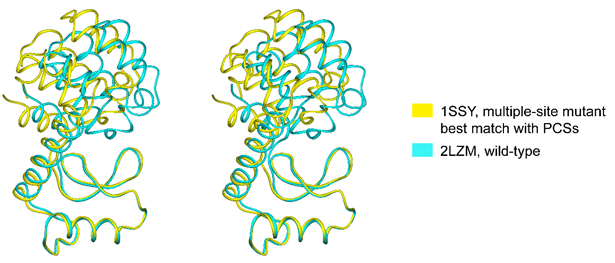
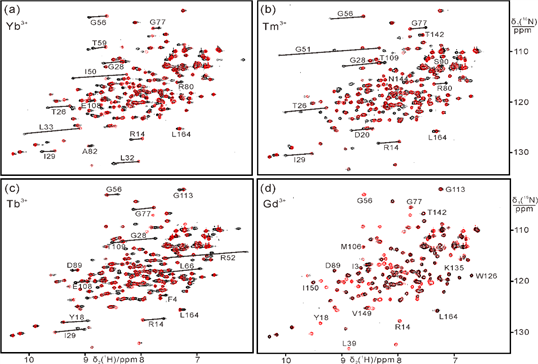
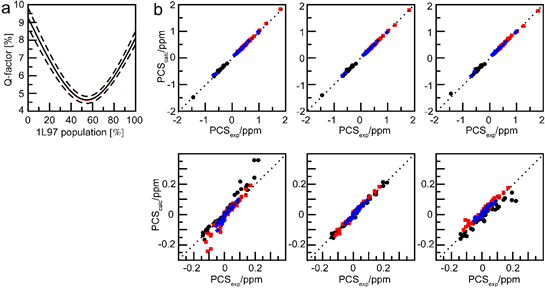
The data indicate that, in solution and in the absence of substrate, the structure of T4 lysozyme is on average more open than suggested by the closed conformation of the crystal structure of wild-type protein.
In collaboration with Professors Gottfried Otting and Thomas Huber at Australian National University.
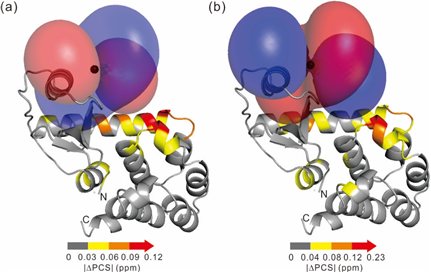
2) Rigid lanthanide binding tag produces narrow distance distributions measured by DEER experiments: NMR and EPR are complementary tools in structure biology (in collaboration with Professor Daniella Goldfarb)
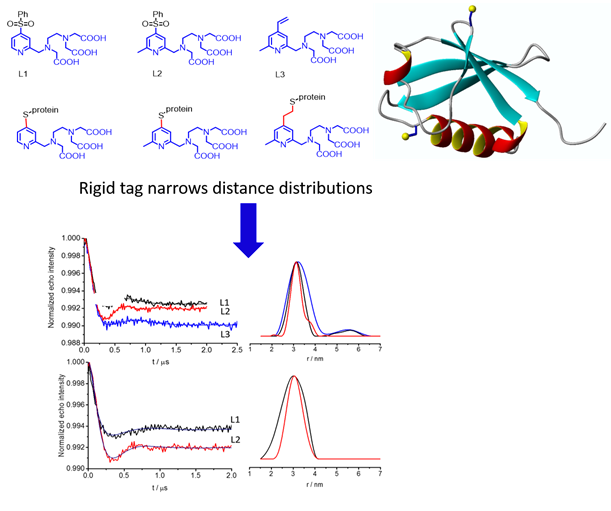
We used NMR spectroscopy optimized paramagnetic tags and conjugated proteins with these rigid tags for DEER measurements (Dalton 2015, 20812).
In collaboration with Professor Daniella Goldfarb at Weizmann Institute of Science, Israel.
Rigid, stable and efficient Gd(III) spin label for in-cell DEER measurements:
In collaboration with Professor Daniella Goldfarb, we reported a new Gd(III) spin label, DO3MA-3BrPy-Gd(III) for in-cell DEER measurements.
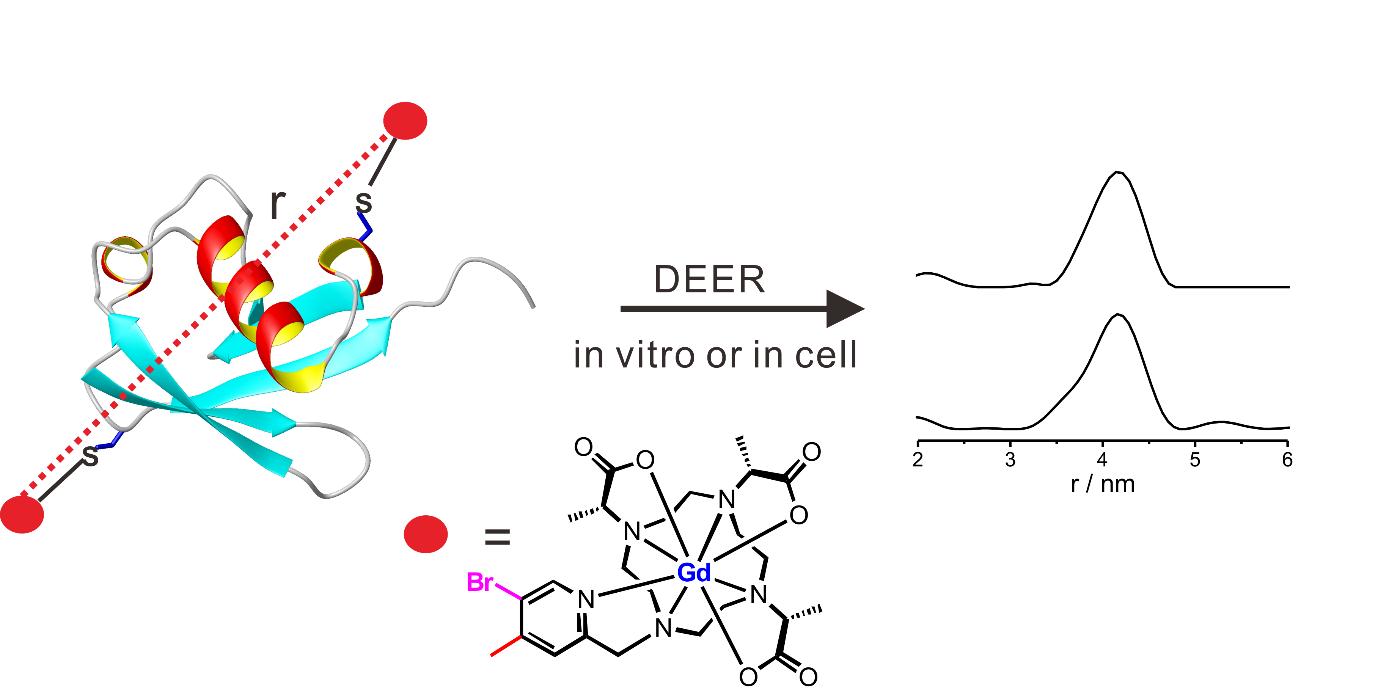
The new Ln(III) spin label has first been optimized by NMR and then applied for PD-EPR. The high performance of DO3MA-3BrPy-Gd(III) is demonstrated on doubly labelled ubiquitin D39C/E64C, both in vitro and in HeLa cells. High-quality DEER data could be obtained in HeLa cells up to 12 h after protein delivery at in-cell protein concentrations as low as 5-10 mM (Angew. Chem. DOI: 10.1002/anie.201611051R1).
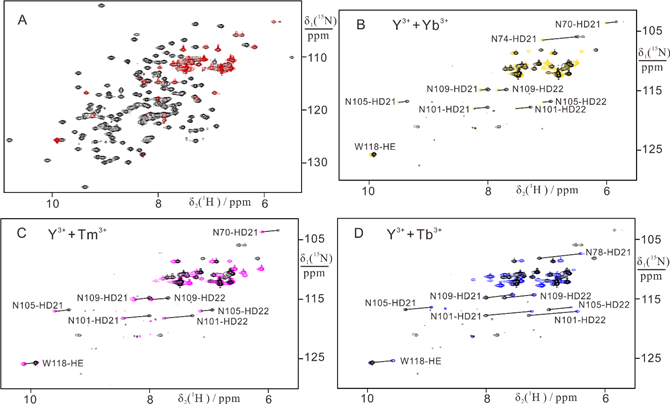
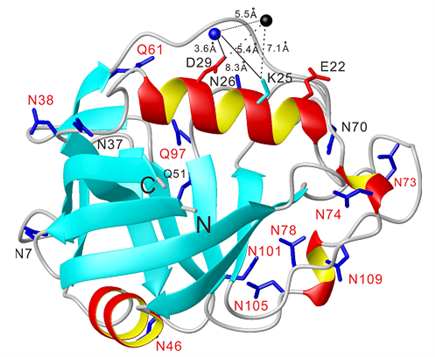
3) In-cell NMR analysis using stable paramagnetic tags: structures and dynamics of proteins delineated by paramagnetic NMR spectroscopy
3D structure determination of a protein in living cells using paramagnetic NMR spectroscopy (in collaboration with Professors Thomas Huber and Conggang Li).
Determining the three-dimensional structure of a protein in living cells remains particularly challenging. We demonstrated that the integration of site-specific tagging proteins and GPS-Rosetta calculations provides a fast and effective way of determining the structures of proteins in living cells, and in principle the interactions and dynamics of protein-ligand complexes (Chem. Commun. 2016, 10237).
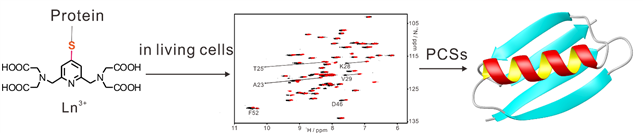
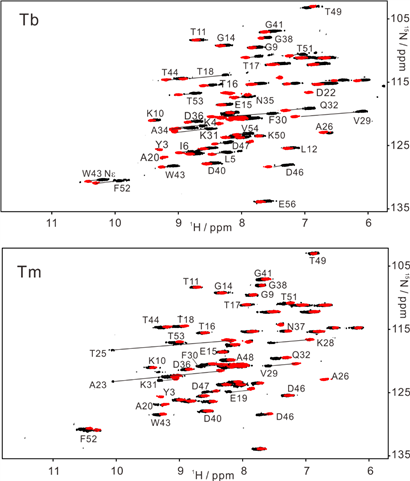
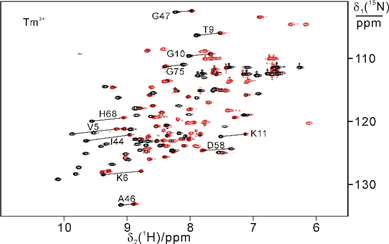
15N-HSQC spectra recorded for GB1 V21C-PyMTA complexed lanthanide ion in living Xenopus laevis oocytes (total acquisition time within 2 hours).
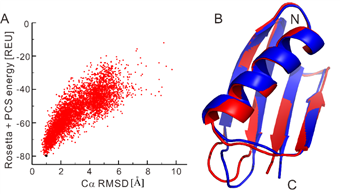
High-resolution structure calculation from in-cell PCS data using GPS-Rosetta. We have presented an efficient way to determine the structure of a protein in living cells by employing paramagnetic restraints from PCSs. PCSs are readily measured by the chemical shift differences observed in 15N-HSQC spectra. The high sensitivity of the experiment allows accurate PCS data to be recorded in living cells where the limited lifetime of the cells under the condition of the NMR measurement prohibits long measurement times and/or protein concentration can be a limiting factor. Moreover, low protein concentration (~0.05 mM) was sufficient for recording 15N-HSQC spectra within 2 hours. This work was highlighted in ChemistryWorld,
https://www.chemistryworld.com/news/molecular-beacon-homes-in-on-protein-structures/1017298.article.
4) 3D structure of transient and unstable enzyme intermediate determined by paramagnetic NMR spectroscopy
Enzyme catalysis of chemical reactions relies on conformational plasticity but structural information on transient intermediates is difficult to obtain. Here we show that the three-dimensional (3D) structure of an unstable, low abundance enzymatic intermediate can be determined by nuclear magnetic resonance (NMR) spectroscopy. The approach is demonstrated for Staphylococcus aureus sortase A (SrtA), which is an established drug target and biotechnological reagent. SrtA is a transpeptidase that converts an amide bond of substrate peptide to a thioester. By measuring pseudocontact shifts (PCS) generated by a site-specific cysteine-reactive paramagnetic tag that does not react with the active site residue Cys184, a sufficient number of restraints could be collected to determine the 3D structure of the unstable thioacyl intermediate of SrtA that is present only as a minor species under non-equilibrium conditions (Angew. Chem. 2016, 55, 13744).
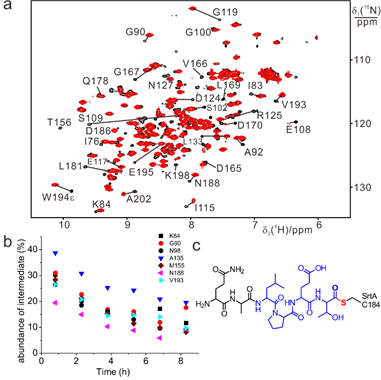
Low abundance of thioester intermediate formed by SrtA and QALPETG peptide determined by NMR.
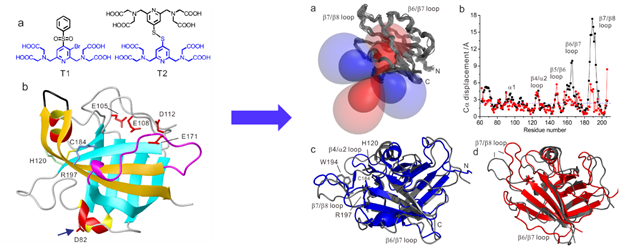
To the best of our knowledge, the structure of the short-lived thioester intermediate of SrtA determined here with the help of PCSs presents the first 3D structural view, in solution, of an enzymatic intermediate present only as a minor species under non-equilibrium conditions. The structure determination relied on lanthanide tags of carefully tuned reactivity to avoid modifying Cys184 at the active site.
5) XIAP and copper homeostasis
The structural properties of BIR1 of XIAP were characterized using high resolution NMR spectroscopy in vitro and in different physiological environments. The effect of copper addition in both oxidation states on BIR1 was then investigated. Wild-type BIR1 exists as a stable homodimer in solution, the dimerization can be impaired by two point mutations at the dimer interface, D71N/R72E, resulting in a well-folded monomeric protein. No interaction between BIR1 and copper(I) was observed both in vitro and in-cell. Remarkably cysteine 12, which resides in a well conserved TCVP motif in the unstructured N-terminal tail among XIAP homologues, presents a very low redox potential (~-300 mV), and is found to react with copper (II) leading to the formation of either a disulfide-linked protein dimer or an adduct with glutathione in vitro and in-cell lysate.
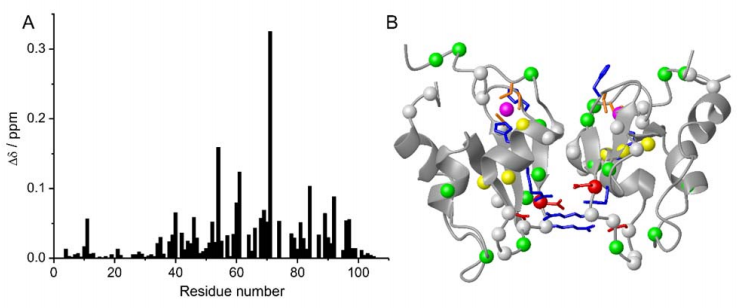
Chemical shift perturbations of D71N/R72E mutant on BIR1. (A) Plot of chemical shift perturbations caused by D71N/R72E mutation on BIR1. The chemical-shift differences were calculated as δ =((ΔδH)2+(ΔδN/10) 2)1/2 . (B) Chemical shift differences between WT BIR1 and D71N/R72E mutant plotted on the dimeric structure of BIR1 (PDB code: 2QRA), of which the Cα atoms are shown in red spheres with δ > 0.2 ppm, yellow spheres with 0.1
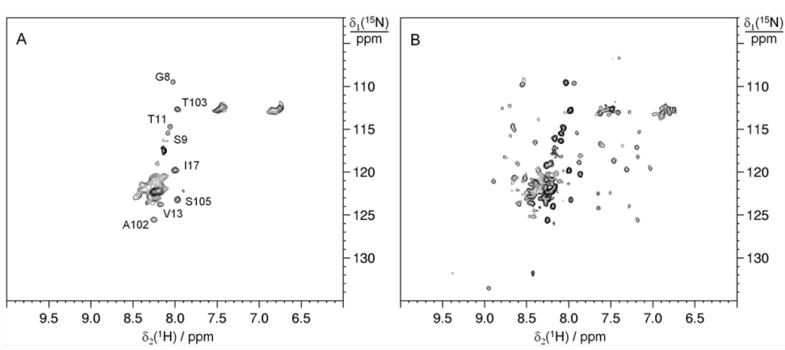
15N SOFAST-HMQC spectra of WT BIR1 in HEK293T cells and cell lysate. A) on a sample of HEK293T cells expressing WT BIR1 and B) on the corresponding cell lysate.
We show that the BIR1 domain of human XIAP forms a homodimer in solution. The dimer is destabilized by removing two salt bridges at the binding interface, resulting in the monomeric D71N/R72E mutant. Cysteine 12 in the TCVP motif is prone to oxidation, resulting in the formation of a disulfide bond between two BIR1 subunits, or in the formation of a mixed BIR1-GSH disulfide bond in a physiological environment. Cu(I) binding occurs non-specifically and with low affinity, therefore is unlikely that BIR1 directly binds copper in a physiological environment. Instead, Cu(II) is able to oxidize C12 both in buffer and in the cell lysate.
Publications:
Hou MM, Polykretis P, Luchinat E, Wang X, Chen SN, Zuo HH, Yang Y, Chen JL, Ye Y, Li C, Banci L, Su XC*. Solution structure and interaction with copper in vitro and in living cells of the first BIR domain of XIAP. Sci. Rep. 2017, 7:16630.
Li X, Wu G, Yang Y, Fu S, Liu X, Kang H, Yang X*, Su XC*, Shen Y*. Calmodulin dissociates the STIM1-Orial complex and STIM1 oligomers. Nat. Commun. 2017, 8: 1042.
Gao J, Liang E, Ma R, Li F, Liu Y, Liu J, Jiang L, Li C, Dai H, Wu J, Su XC, He W*, Ruan K*. Fluorine pseudocontact shifts used for characterizing the protein-ligand interaction mode in the limit of NMR intermediate exchange. Angew. Chem. Int. Ed. Engl. 2017, 56, 12982-12986.
Yang Y, Gong YJ, Litinov A, Liu HK, Yang F, Su XC*, Goldfarb D*. Generic Tags for Mn(II) and Gd(III) spin labels for distance measurements in proteins. Phys. Chem. Chem. Phys. 2017, 19, 26944-26956.
Cao C, Wang S, Cui T, Su XC*, Chou J*. Ion and inhibitor binding of the double-ring ion selectivity filter of the Mitochondrial calcium uniporter. Proc. Natl. Acad. Sci. USA, 2017, 114, E2846-E2851.
Yang Y, Yang F, Gong YJ, Chen Jl., Goldfarb D*, Su XC*. A reactive, rigid Gd(III) labelling tag for in-cell EPR distance measurements in proteins. Angew. Chem. Int. Ed. Engl. 2017, 56, 2914-2918.
Ma FH, Wang X, Chen JL, Wen X, Sun H*, Su XC*. Deciphering the multisite interactions of a protein and its ligand at atomic resolution by using sensitive paramagnetic effects. Chem. Eur. J. 2017, 23, 926-934.
Li ML, Yang S, Su XC, Wu HL, Yang LL, Zhu SF, Zhou QL*. Mechanism Studies of Ir-Catalyzed Asymmetric Hydrogenation of Unsaturated Carboxylic Acids. J. Am. Chem. Soc. 2017, 139, 541-547.
Chen JL, Wang X, Yang F, Cao C, Otting G, Su XC*. 3D Structure determination of an unstable transient enzyme intermediate by paramagnetic NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2016, 55, 13744-13748.
Yang F, Wang X, Pan BB, Su XC*. Single-armed phenylsulfonated pyridine derivative of DOTA is rigid and stable paramagnetic tag in protein analysis. Chem. Commun. 2016, 52, 11535-11538.
Pan BB, Yang F, Ye Y, Wang JT, Li C*, Huber T*, Su XC*. Structural determination of proteins in living cells by paramagnetic NMR spectroscopy. Chem. Commun. 2016, 52, 10237-10240.
Ma RS, Li QF, Wang AD, Zhang JH, Liu ZJ, Wu JH, Su XC*, Ruan K*. Determination of pseudocontact shifts of low-populated excited states by NMR chemical exchange saturation transfer. Phys. Chem. Chem. Phys. 2016, 18, 13794-13798.
Yang Y, Huang F, Huber T, Su XC*. Site-specific tagging proteins with a rigid, small and stable transition metal chelator, 8-hydroxyquinoline, for paramagnetic NMR analysis. J. Biolmol. NMR 2016, 64, 102-113.
Chen JL, Yang Y, Zhang LL, Liang H, Huber T*, Su XC*, Otting G*. Analysis of the solution conformations of T4 lysozyme by paramagnetic NMR spectroscopy. Phys. Chem. Chem. Phys. 2016, 18, 5850-5859.
Martorana A, Yang Y, Zhao Y, Li QF, Su XC*, Goldfarb D*. Mn(II) tags for DEER distance measurements in proteins via C-S attachment. Dalton 2015, 44, 20812-20816.
Yang Y, Wang JT, Pei YY, Su XC*. Site-specific tagging proteins via a rigid, stable and short thiolether tether for paramagnetic spectroscopic analysis. Chem. Commun. 2015, 51, 2824-2827.
Wang Z, Yang X, Guo S, Yang Y, Su XC, Shen Y, Long J. Crystal structure of the ubiquitin-like domain-CUT repeat-like tandem of special AT-rich sequence binding protein 1 (SATB1) reveals a coordinating DNA-binding mechanism. J. Biol. Chem. 2014, 289, 27376-27385.
Cao C, Chen JL, Yang Y, Huang F, Otting G, Su XC*. Selective 15N-labeling of the side-chain amide groups of asparagine and glutamine for applications in paramagnetic NMR spectroscopy. J. Biomol. NMR. 2014, 59, 251-261.
Ma FH, Chen JL, Li QF, Su XC*. Kinetic assay of Michael addition like thiol-ene reaction and the insight into protein bioconjugation. Chem. Asian J. 2014, 9, 1808-1816.
Yang Y, Chen JL, Su XC*. PCS in structural biology. Chin. J. Magn. Reson. (杨茵,陈家良,苏循成*. 蛋白质顺磁标记技术与生物核磁共振中的赝接触位移,波谱学杂志) 2014, 31, 155-171.
Su XC, Wang Y, Yagi H, Shishmarev D, Mason CE, Smith PJ, Vandevenne M, Dixon NE, Otting G*. Bound or free: interaction of the C-terminal domain of Escherichia coli single-stranded DNA-binding protein (SSB) with the tetrameric core of SSB. Biochemistry 2014, 52, 1925-1934.


2010 年第一届研究生合影

2013年6月第一届研究生答辩

2013年全体研究生合影


2014年研究生毕业照


2015年研究生毕业照




2016年研究生毕业照

2017年课题组合影
